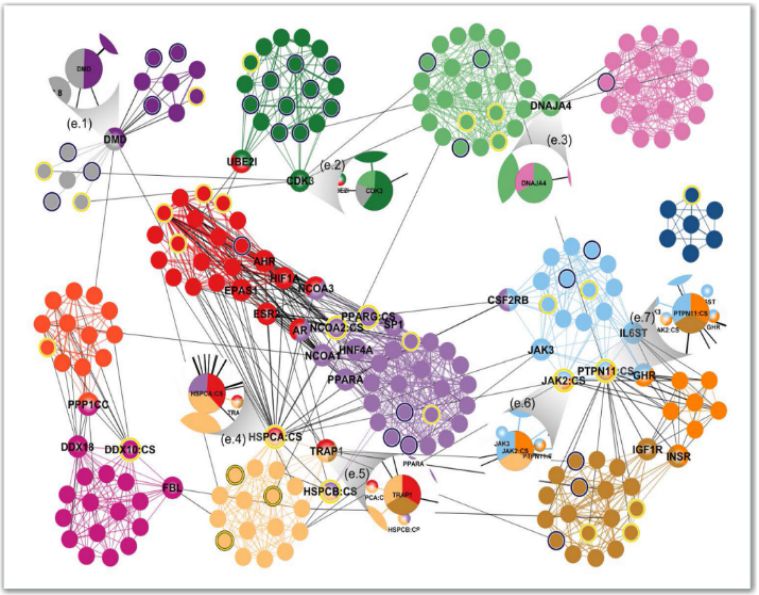
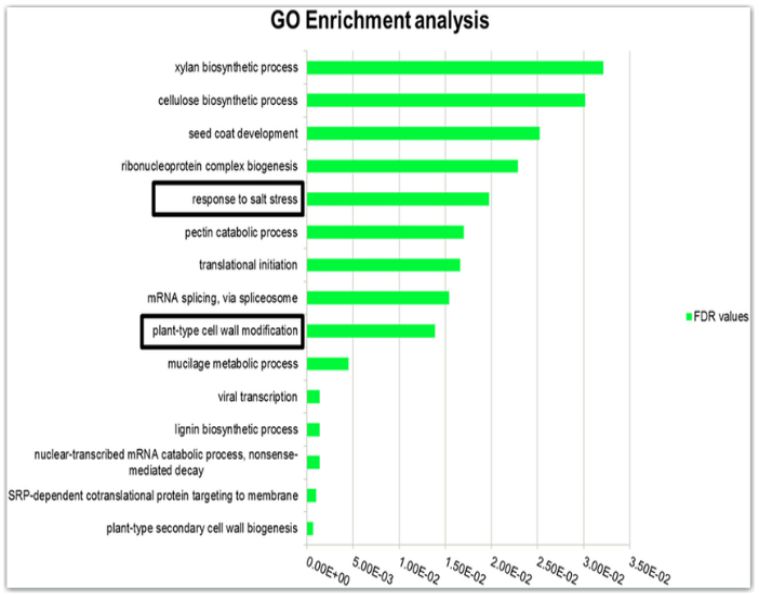

Welcome to the GWASdmp
In recent years, genome-wide association studies (GWAS) have attracted great attention for their ability to detect disease-associated loci of multifactorial disorders. However, the interpretation of these genetic variants still remains difficult. GWASdmp is an online integration tool for GWAS data mining in the Post-GWAS Era. It comprises five modules, including SNP analysis, causal gene prediction, functional analysis, gut microbiota and environmental exposure associations analysis.
In GWASdmp, users can:
✔ Run SNP analysis, such as localization, expansion, annotation;
✔ Predict causal genes, such as eQTL analysis, gene-gene interaction analysis and
protein-protein interaction analysis;
✔ Conduct function analysis, such as function prediction and gene set enrichment analysis;
✔ Explore the association between gut microbiota and complex disorders;
✔ Explore the association between environmental chemical exposures and complex disorders.
Server Monitor

GWASdmp Summary
Welcome to the GWASdmp
2020-12-22: GWASdmp Web Server 1.0 Releasing Publicly!
About Us
Fanglin Guan, Health Science Center, Xi’an Jiaotong University, 76 Yanta West Road, Xi’an, China;
Email: fanglingguan@163.com
Shiqiang Cheng, Health Science Center, Xi’an Jiaotong University, 76 Yanta West Road, Xi’an, China;
Email: chengsq0701@stu.xjtu.edu.cn
Feng Zhang, Health Science Center, Xi’an Jiaotong University, 76 Yanta West Road, Xi’an, China;
Email: fzhxjtu@mail.xjtu.edu.cn
Visit Statistics